Next-Generation Sequencing for Uveal Melanoma
Identifying somatic mutations in seven important genes relevant to uveal melanoma
The test includes hotspot mutations in the genes GNAQ, GNA11, CYSLTR2, PLCB4, and SF3B1, mutations in exons 1-2 of EIF1AX, and all coding exons of the BAP1 gene.
This information, together with results from the DecisionDx®-UM and DecisionDx®-PRAME gene expression profile (GEP) tests, can help to build a comprehensive genomic profile of an individual UM tumor from a single biopsy. The genomic information can be used to inform patient care and to become more useful in the future as research and therapeutics evolve.

DecisionDx-UMSeq can be ordered for patients who are having DecisionDx-UM GEP testing performed. Castle Biosciences has optimized the sequencing test so that it can be run using the same fine-needle aspiration biopsy (FNAB) tissue specimen submitted for GEP testing.
Mutations in four genes affect G-protein-coupled receptor signaling
In UM, mutations in GNAQ, GNA11, CYSLTR2, or PLCB4 result in constitutive activation of G-protein-coupled receptor signaling pathways, such as MAPK, PI3K, PKC, Hippo, etc. (Van Raamsdonk, 2009; 2011; Johansson, 2016; Moore, 2016).
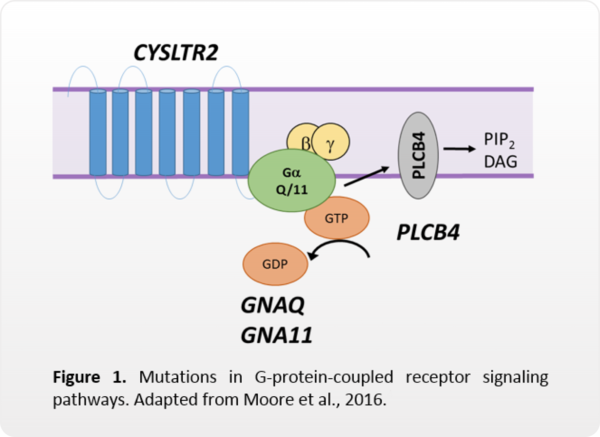
While some of these pathways may be targeted with inhibitory therapies, these mutations are not currently markers for drug response. These mutations are also not known to be prognostic.
The majority of UM tumors will have a mutation in GNAQ or GNA11, while CYSLTR2 and PLCB4 mutations are less common. These mutations are usually mutually exclusive. The presence of one of these mutations may provide some confidence that a melanocytic tumor was sampled. However, other eye lesions can have these mutations, and some UM tumors will not have a mutation in any of these four genes. These mutations should not be used exclusively to rule-in or rule-out a UM diagnosis in the absence of other clinical/pathological features.
Learn more about gene mutations
The GNAQ gene encodes for the alpha subunit of the heterotrimeric G protein complex that couples to seven transmembrane domain receptors (G-protein coupled receptors, GPCRs) to affect intracellular signaling.
The alpha subunit has intrinsic hydrolase activity that regulates the availability of active GTP for GPCRs, influencing downstream signaling pathways, including MAPK, PKC, PI3K and Hippo. Mutations in GNAQ primarily affect two amino acids: Q209 (exon 5) or R183 (exon 4)(Van Raamsdonk, 2009).
Both amino acid changes curtail the subunit’s GTPase activity and inhibit the conversion of GTP to inactive GDP. This results in the constitutive activation of the pathways downstream of the GPCR.
Drugs targeting these various pathways are being explored as therapeutics for uveal melanoma(Shoushtari & Carvajal, 2014). Mutations in GNAQ occur in approximately 40-45% of uveal melanomas (Van Raamsdonk, 2009; Van Raamsdonk, 2010; Onken, 2008; Decatur, 2016) and are also found in blue nevi, cutaneous melanomas, mucosal melanomas, and other lesions. GNAQ is highly homologous to GNA11, and mutations in these genes are usually mutually exclusive with each other, as well as with CYSLTR2 and PLCB4(Moore, 2016; Johansson, 2016).
Approximately 85-90% of uveal melanomas will have a mutation in either GNAQ or GNA11(Van Raamsdonk, 2010). GNAQ mutations are not associated with prognosis in uveal melanoma.
The GNA11 gene encodes for the alpha subunit of the heterotrimeric G protein complex that couples to seven transmembrane domain receptors (G-protein coupled receptors, GPCRs) to affect intracellular signaling.
The alpha subunit has intrinsic hydrolase activity that regulates the availability of active GTP for GPCRs, influencing downstream signaling pathways, including MAPK, PKC, PI3K and Hippo. Mutations in GNA11 primarily affect two amino acids: Q209 (exon 5) or R183 (exon 4)(Van Raamsdonk, 2010).
Both amino acid changes curtail the subunit’s GTPase activity and inhibit the conversion of GTP to inactive GDP. This results in the constitutive activation of the pathways downstream of the GPCR.
Drugs targeting these pathways are being explored as therapeutics for uveal melanoma(Shoushtari & Carvajal, 2014). Mutations in GNA11 occur in approximately 40%-45% of uveal melanomas(Van Raamsdonk, 2010; Decatur, 2016), and are also found in other lesions, including blue nevi, cutaneous melanomas, and mucosal melanomas.
GNA11 is highly homologous to GNAQ, and mutations in these genes are usually mutually exclusive with each other, as well as with CYSLTR2 and PLCB4(Moore, 2016; Johansson, 2016).
Approximately 85-90% of uveal melanomas will have a mutation in either GNAQ or GNA11 (Van Raamsdonk, 2010). GNA11 mutations are not associated with prognosis in uveal melanoma.
The CYSLTR2 gene encodes for a G-protein coupled receptor in the rhodopsin-like family called cysteinyl leukotriene receptor 2. The receptor is stimulated by leukotrienes to mediate downstream signaling and has been shown to activate the Gaq subunit.
A hotspot mutation in CYSLTR2 has been identified in uveal melanoma that results in an amino acid change at a L129(Moore, 2016). This mutation results in constitutive activation of the receptor through the Gaq subunit and may activate downstream pathways similar to those activated by GNAQ and GNA11 mutations in uveal melanoma, including the MAPK, PKC, PI3K, and Hippo pathways.
CYSLTR2 mutations have been shown to occur in approximately 3% of uveal melanomas and are mutually exclusive with GNAQ, GNA11, and PLCB4 mutations (Moore, 2016). The CYSLTR2 mutation is not associated with prognosis in uveal melanoma.
The PLCB4 gene encodes for phospholipase C, beta 4, which is a downstream target of the Gaq and Ga11 subunits of G-protein-coupled receptors. PLCB4 catalyzes the formation of inositol 1,4,5-triphosphate (IP3) and diacylglycerol (DAC) from phosphatidylinositol 4,5-bisphosphate (PIP2) and thus plays a role in calcium-mediated intracellular signaling.
A hotspot mutation resulting in an amino acid change at D630 in PLCB4 is thought to disrupt the catalytic core of PLCB4 and result in aberrant signaling downstream of the Ga subunits (Johansson, 2016).
PLCB4 mutations occur in 4-7% of uveal melanomas and are mutually exclusive with mutations in GNAQ, GNA11, and CYSLTR2 (Moore, 2016; Johansson, 2016) PLCB4 mutations also occur in cutaneous melanoma but at different genetic locations than in uveal melanoma. The PLCB4 mutation is not associated with prognosis in uveal melanoma.
Mutations in three genes are associated with differential clinical outcomes
Mutations occurring in EIF1AX, SF3B1, and BAP1 are usually mutually exclusive
It is well known that mutations in BAP1 in UM are associated with a higher risk of metastasis and there is considerable overlap with a Class 2 GEP (Harbour, 2010). Some BAP1 mutations are germline and can be inherited (Abdel-Rahman, 2011; Gupta, 2015), but the majority are somatic and thus confined to the tumor.
In retrospective studies, having an EIF1AX or SF3B1 mutation has been shown to be associated with a better prognosis than having a BAP1 mutation (Martin, 2013; Ewens, 2014; Decatur, 2016; Furney, 2013; Harbour, 2013; Yavuzyigitoglu, 2016).
Tumors with EIF1AX mutations may have the lowest risk of metastasis compared to those with SF3B1 mutations, which may be prone to late metastasis.


Importantly, in a recent retrospective study comparing the prognostic value of these 3 mutations when using the DecisionDx-UM GEP test, a Class 2 result was the only significant, independent predictor of metastasis and UM-related mortality in multivariate analysis (Decatur, 2016). Therefore, the GEP test currently provides the best prognostic information about the tumor.
The EIF1AX gene encodes for the eukaryotic translation initiation factor 1A, x-linked protein. This protein plays a role in the initiation of translation of mRNA to protein. In uveal melanoma, mutations in EIF1AX have been reported in the first two exons of the gene, which encode for the unstructured N-terminal tail of the EIF1AX protein (Martin, 2013).
EIF1AX mutations occur in approximately 17-24% of uveal melanomas and are usually not found in conjunction with SF3B1 or BAP1 mutations (Decatur, 2016). In small, retrospective, single center studies, mutations in EIF1AX have been associated with a good prognosis in uveal melanoma (Ewens, 2014, Yavuzyigitoglu, 2016).
The SF3B1 gene encodes for splicing factor B3 subunit 1. This protein is a component of the U2 small nuclear ribonucleoprotein complex and plays a role in splicing of pre-mRNA.
In uveal melanoma, SF3B1 mutations usually result in an amino acid change at R625 (Harbour, 2013; Martin, 2013) that may result in differential splicing of protein coding genes (Furney, 2013). SF3B1 mutations occur in 18-24% of uveal melanomas (Harbour, 2013; Furney, 2013, Decatur, 2016), and can also occur in other malignancies, including breast cancer, pancreatic cancer, chronic lymphocytic leukemia, and myelodysplastic syndrome.
Mutations in SF3B1 are usually not found in conjunction with EIF1AX or BAP1 mutations. In small, retrospective, single center studies, SF3B1 mutations have been associated with a favorable prognosis but may be linked to a risk for late metastasis (Yavuzyigitoglu, 2016; Harbour, 2013).
The BAP1 gene is a 17-exon gene located on chromosome 3 and encodes for BRCA1-associated protein 1, a ubiquitin carboxy-terminal hydrolase (deubiquitinating enzyme) that can interact with multiple factors, including BRCA1, BARD1, ASXL1, and HCFC1, and functions as a tumor suppressor. BAP1 mutations can occur anywhere along the gene and generally result in loss of expression and/or functional inactivation. BAP1 mutations occur in approximately 40-45% of uveal melanomas (Harbour, 2010; Decatur, 2016) and are not usually found in conjunction with SF3B1 or EIF1AX mutations. In uveal melanoma, retrospective studies have shown that BAP1 mutations are associated with an increased risk of metastasis (Decatur, 2016) and are strongly correlated with other unfavorable prognostic characteristics, such as a Class 2 gene expression profile and monosomy 3 (Harbour, 2010; Ewens, 2014).
BAP1 mutations can be somatic, meaning they arose spontaneously in the tumor, or germline, meaning they are present in every cell in the body and can be heritable. Most BAP1 mutations in uveal melanoma are somatic in origin, and only a very small percentage (2-5%) of uveal melanoma tumors will have a germline BAP1 mutation (Aoude 2013; Gupta 2015). Germline BAP1 mutations are associated with BAP1 tumor predisposition syndrome and patients with this syndrome may be at an elevated risk for uveal melanoma, cutaneous melanoma, mesothelioma, renal cell carcinoma, and other conditions (Abdel-Rahman, 2011; Njauw, 2012; Gupta, 2015; Rai, 2016).
Understanding the DecisionDx-UMSeq results
The DecisionDx-UMSeq test results can be expected in 2-4 weeks after receipt of the sample in our laboratory.
The DecisionDx-UMSeq report will tell you if clinically relevant mutations (variants) were found in any of the seven gene targets. For each mutation found, the report will describe:
- Genomic location of the mutation (where in the gene it occurred)
- Type of mutation (eg, missense, nonsense)
- Functional change that occurs because of the mutation (ie, an amino acid change in the protein)
- Frequency that the mutation was detected in the sample (variant allele frequency)
- Potential consequences of that mutation on gene function, as well as relevant literature references
The Tier and Level of Evidence will also be described for each mutation, as recommended by the College of American Pathologists (CAP), the American Society of Clinical Oncology (ASCO), and the Association for Molecular Pathologists (AMP)(Li, 2017).
Ordering DecisionDx-UMSeq
BAP1 germline testing information
If your patient has a detected BAP1 mutation and is interested in germline testing, Castle Biosciences can provide information to assist with finding testing services and genetic counseling. Please call Castle Biosciences at 866-788-9007, option 1 for these resources.
References
- Abdel-Rahman MH, Pilarski R, Cebulla CM, et al. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J Med Genet 2011;48:856-9.
-
Aoude LG, Vajdic CM, Kricker A, et al. Prevalence of germline BAP1 mutation in a population-based sample of uveal melanoma cases. Pigment Cell Melanoma Res 2013;26:278-9.
-
Decatur CL, Ong E, Garg N, et al. Driver mutations in uveal melanoma: associations with gene expression profile and patient outcomes. JAMA Ophthalmol 2016;134:728-33.
-
Ewens KG, Kanetsky PA, Richards-Yutz J, et al. Chromosome 3 status combined with BAP1 and EIF1AX mutation profiles are associated with metastasis in uveal melanoma. Invest Ophthalmol Vis Sci 2014;55:5160-7.
-
Furney SJ, Pedersen M, Gentien D, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov 2013;3:1122-9.
-
Gupta MP, Lane AM, DeAngelis MM, et al. Clinical characteristics of uveal melanoma in patients with germline BAP1 mutations. JAMA Ophthalmol 2015;133:881-7.
-
Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010;330:1410-3.
-
Harbour JW, Roberson ED, Anbunathan H, et al. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet 2013;45:133-5.
-
Johansson P, Aoude LG, Wadt K, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 2016;7:4624-31.
-
Martin M, Masshofer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet 2013;45:933-6.
-
Moore AR, Ceraudo E, Sher JJ, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet 2016;48:675-80.
-
Njauw CN, Kim I, Piris A, et al. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PLoS One 2012;7: e35295.
-
Onken MD, Worley LA, Long MD, et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci 2008;49:5230-4.
-
Rai K, Pilarski R, Cebulla CM, et al. Comprehensive review of BAP1 tumor predisposition syndrome with report of two new cases. Clin Genet 2016;89:285-94.
-
Shoushtari AN and Carvajal RD. GNAQ and GNA11 mutations in uveal melanoma. Melanoma Res 2014;24:525-34.
-
Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 2009;457:599-602.
-
Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med 2010;363:2191-9.
-
Yavuzyigitoglu S, Koopmans AE, Verdijk RM, et al. Uveal melanomas with SF3B1 Mutations: a distinct subclass associated with late-onset metastases. Ophthalmology 2016;123:1118-28.



