
Learn about the testing process for early skin cancer detection
Using qRT-PCR technology, MyPath Melanoma objectively distinguishes melanoma from benign nevi with greater than 90% accuracy in three independent clinical validation studies. In the initial discovery phase of test development, candidate genes were selected based upon published data demonstrating their differential expression in melanoma compared with benign nevi or their increased expression in aggressive tumors. This panel was refined to most effectively differentiate benign from malignant melanocytic lesion in a training cohort of archival formalin-fixed paraffin-embedded lesions (n=464).
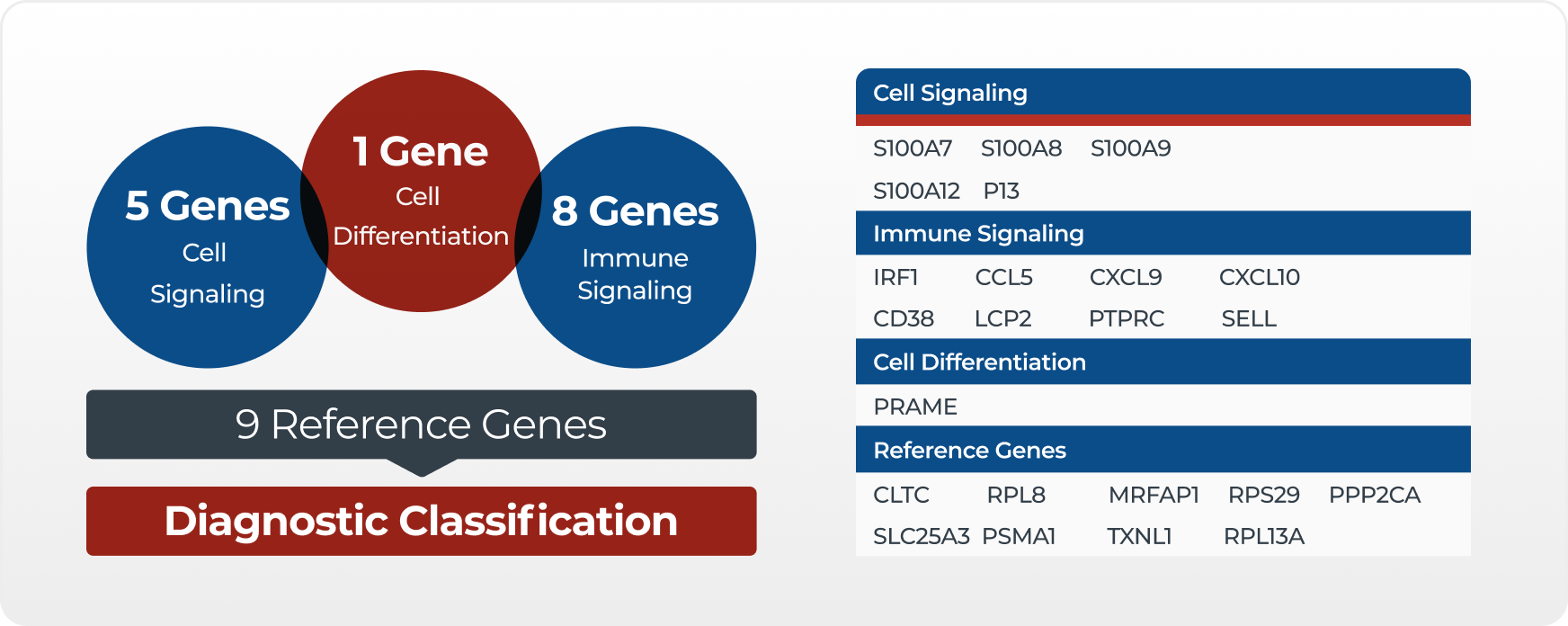
Statistical modeling identified a subset of 14 genes, grouped into three distinct gene components, which provided the greatest sensitivity and specificity. These 14 signature genes are involved in cell differentiation and cellular immune signaling. Expression of the signature genes is normalized to that of nine reference genes prior to the application of a weighted algorithm, which combines the measurements of all signature genes and generates a single numerical score.
MyPath Melanoma testing process
MyPath Melanoma is performed in Castle Biosciences’ CLIA-certified, CAP-accredited and NY state approved laboratory using formalin-fixed, paraffin embedded (FFPE) primary tumor tissue from either a biopsy or excision.

Tissue biopsy submitted
Tissue blocks or slides from the primary tumor tissue biopsy are sent to our CLIA certified, CAP-accredited and NY state approved laboratory for testing.

RNA/DNA preparation
Once tissue is received in our CLIA certified, CAP-accredited, & NY state approved laboratory, RNA is isolated from the tissue and converted to cDNA. The cDNA for each of the 23 genes is amplified and the gene signature recorded.

Signature interpretation
Using a proprietary modeling algorithm, the individual patient's gene expression signature is assessed.

MyPath Melanoma reporting
The comprehensive report provides the MyPath Melanoma score and its associated categorization of suggestive of benign, intermediate, or suggestive of malignant.
MyPath Melanoma gene component information

The first component is two measurements of the gene PRAME, which stands for preferentially expressed antigen in melanoma. PRAME encodes a cancer-testis protein, which is aberrantly expressed in melanoma. It appears to contribute to tumorigenesis by functioning as a dominant repressor of retinoic acid receptor signaling and/or down-regulation of TRAIL expression.

The second component contains five genes from the S100A family: S100A7, S100A8, S100A9, S100A12, and PI3. The products of these genes are involved in multiple cellular processes. S100A9 is a calcium-binding protein, often found in combination with S100A8 as part of an immunogenic protein heterodimer. Increased S100A8 and S100A9 levels are detected in many malignant neoplasms, both within tumor cells and within infiltrating immune cells.

The third component contains eight genes involved in tumor immune response signaling: CCL5, CD38, CXCL10, CXCL9, IRF1, LCP2, PTPRC, and SELL. Many of these genes produce chemokines or chemokine receptors that regulate leukocyte trafficking. Chemokines can suppress or promote the growth of a neoplasm by acting on cells of the tumor microenvironment, including leukocytes, endothelial cells, and fibroblasts. But, they may also affect tumor cells themselves by regulating migration, invasion, proliferation, and resistance to chemotherapy.

The fourth component is a group of nine housekeeping genes whose measurement allows normalization of the RNA expression for analysis.
Get started with MyPath Melanoma






